Note
Click here to download the full example code
A quick example¶
Amputation is the opposite of imputation: the generation of missing values in complete datasets. That is useful in an experimental setting where you want to evaluate the effect of missing values on the outcome of a model.
MultivariateAmputation is designed following scikit-learn’s fit and transform paradigm, and can therefore seamless be integrated in a larger data processing pipeline.
Here, we give a short demonstration. A more extensive example can be found in this example. For people who are familiar with the implementation of multivariate amputation in R-function ampute, this blogpost gives an overview of the similarities and differences with MultivariateAmputation. Inspection of an incomplete dataset can be done with mdPatterns.
Note that the amputation methodology itself is proposed in Generating missing values for simulation purposes and in The dance of the mechanisms.
# Author: Rianne Schouten <https://rianneschouten.github.io/>
# Co-Author: Davina Zamanzadeh <https://davinaz.me/>
Transforming one dataset¶
Multivariate amputation of one dataset can directly be performed with
fit_transform. Inspection of an incomplete dataset can be done withmdPatterns. By default,MultivariateAmputationgenerates 1 pattern with MAR missingness in 50% of the data rows for 50% of the variables.
import numpy as np
from pyampute.ampute import MultivariateAmputation
from pyampute.exploration.md_patterns import mdPatterns
seed = 2022
rng = np.random.default_rng(seed)
m = 1000
n = 10
X_compl = rng.standard_normal((m, n))
ma = MultivariateAmputation(seed=seed)
X_incompl = ma.fit_transform(X_compl)
mdp = mdPatterns()
patterns = mdp.get_patterns(X_incompl)
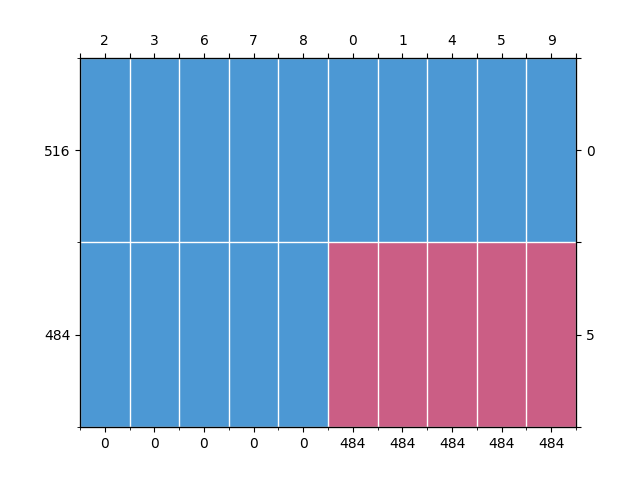
Out:
/home/dav/research/pyampute/pyampute/exploration/md_patterns.py:120: FutureWarning: The frame.append method is deprecated and will be removed from pandas in a future version. Use pandas.concat instead.
group_values = group_values.append(colsums, ignore_index=True)
A separate fit and transform¶
Integration in a larger pipeline requires separate
fitandtransformfunctionality.
from sklearn.model_selection import train_test_split
X_compl_train, X_compl_test = train_test_split(X_compl, random_state=2022)
ma = MultivariateAmputation()
ma.fit(X_compl_train)
X_incompl_test = ma.transform(X_compl_test)
Integration in a pipeline¶
A short pipeline may look as follows.
from sklearn.pipeline import make_pipeline
from sklearn.impute import SimpleImputer
import matplotlib.pyplot as plt
pipe = make_pipeline(MultivariateAmputation(), SimpleImputer())
pipe.fit(X_compl_train)
X_imp_test = pipe.transform(X_compl_test)
By default, SimpleImputer imputes with the mean of the observed data. It is therefore like that we find the median in 50% of the rows (of the test set, which contains 25% of m) for 50% of the variables.
medians = np.nanmedian(X_imp_test, axis=0)
print(np.sum(X_imp_test == medians[None, :], axis=0))
Out:
[ 0 0 120 120 120 0 120 120 0 0]
For more information about pyampute’s parameters, see A mapping from R-function ampute to pyampute. To learn how to design a more thorough experiment, see Evaluating missing values with grid search and a pipeline.
Total running time of the script: ( 0 minutes 0.226 seconds)